Master Practical Training Project/ Master’s thesis
Supervision:
- Roxane Licandro, Medical University of Vienna, Department of Biomedical Imaging and Image-guided Therapy, Computational Imaging Research, roxane.licandro@meduniwien.ac.at
Cooperation Partner:
- Medical University of Vienna Department of Neurosurgery and Department of Biomedical Imaging and Image-guided Therapy – Division of General and Paediatric Radiology
Problem Statement
Neurosurgical interventions are carefully planned procedures, where structural and functional Magnetic Resonance Imaging data before (prae), during (intra), and after (post) surgery are acquired to monitor the procedure and document the outcome of a patient. For performing longitudinal analysis, the alignment of these acquisitions is a key element, however state of the art non-linear registration approaches fail, due to the following challenges:
- the images are acquired using different scanners, with different resolution levels and imaging protocols.
- Beside the patient specific variances of brain shapes, in intra-operative acquisitions unnatural large deformations of brain tissue are observable due to the opening of the skull during the surgical interventions.
- removed brain parts in post-surgical scans can not be considered to establish correspondence in prae-neurosurgical scans, which requires the development of robust alignment strategies.
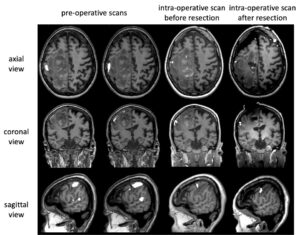
Figure 1: Longitudinal functional MRI preoperative scans (column 1 and 2) and intra-operative scans before the resection of a tumor (column 3) and after (column 4). Axial, coronal, and sagittal views of a single patient’s acquisitions are visualized. Image courtesy Medical University of Vienna.
Goal
The aim of this practical training project/master’s thesis is to develop a patient specific longitudinal alignment strategy between prae-, intra- and post-neurosurgical MRI scans for functional connectivity analysis. Deformable registration and machine learning will be used in combination with robust alignment strategies. Existing diffeomorphic registration frameworks will be extended to fit this challenging application and evaluated against baseline approaches (Voxelmorph [1], SPM [2], LDDMM [3]).
Workflow
- Literature research on existing registration approaches, specifically in the field of non-rigid diffeomorphic deformations with focus on brain alignment.
- Get familiar with MRI/fMRI data (imaging protocols, reading routines, artefacts, etc.)
- Development of a longitudinal alignment pipeline in Python, Tensorflow/Keras or PyTorch.
- Evaluation of the system proposed against baseline approaches.
- Written report/thesis (in English) and final presentation.
For more information please contact Roxane Licandro roxane.licandro@meduniwien.ac.at
References
[1] G. Balakrishnan, A. Zhao, M. R. Sabuncu, J. Guttag and A. V. Dalca, “VoxelMorph: A Learning Framework for Deformable Medical Image Registration,” in IEEE Transactions on Medical Imaging, vol. 38, no. 8, pp. 1788-1800, Aug. 2019, doi: 10.1109/TMI.2019.2897538.
https://github.com/voxelmorph/voxelmorph
[2] J. Ashburner, Computational anatomy with the SPM software, Magnetic Resonance Imaging, Volume 27, Issue 8, 2009, Pages 1163-1174, ISSN 0730-725X, https://doi.org/10.1016/j.mri.2009.01.006.
https://www.fil.ion.ucl.ac.uk/spm/
[3] Beg, M.F., Miller, M.I., Trouvé, A. et al. Computing Large Deformation Metric Mappings via Geodesic Flows of Diffeomorphisms. Int J Comput Vision 61, 139–157 (2005). https://doi.org/10.1023/B:VISI.0000043755.93987.aa
[4] Optimized brain extraction toolbox https://montilab.psych.ucla.edu/fmri-wiki/optibet/
