Status of Master Thesis: open
Supervisors: Paolo Rota and Martin Kampel
(With a positive development of the work there is the possibility of research funding.)
Problem Statement
Despite various suggested approaches towards automated FCM-data analysis, there is no ready-to-use analysis tool available yet, which implements the complete FCM-data analysis pipeline, as described by Bashashati et al. (2009). Such a comprehensive tool would contain automated gating features along with data quality control, normalization, outlier removal and interpretation tasks. Hence, in the presented project, we plan to develop and practically validate such new diagnostic methodology for the clinical practice in patients with AML (see also FlowCLUSTER project website).
Identifying biologically meaningful cell sub-populations is essentially a clustering problem, however, standard clustering methods are impracticable, because size, shape and location of corresponding clusters may vary strongly between samples due to phenotypic differences, inter-laboratory variations, and variations which are inherent in the data acquisition process.
The following picture shows the hierarchical gating structure (selection of different type of cells based on the biological markers – staining) which is used in the context of AML:
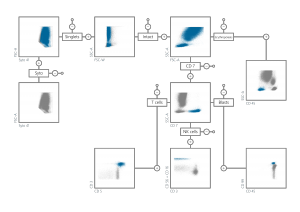 Within the sister project AutoFLOW an automated gating procedure for Acute Lymphblastic Leukeamia (ALL) is being developed (see AutoFLOW). The difference to AML is on the one hand the used staining (markers) and the “biological variation”. The challenge within the automated gating is to detect even small leukaemic populations (~10-20 leukaemic cells) among 300.000 up to 1.000.000 cells.
Within the sister project AutoFLOW an automated gating procedure for Acute Lymphblastic Leukeamia (ALL) is being developed (see AutoFLOW). The difference to AML is on the one hand the used staining (markers) and the “biological variation”. The challenge within the automated gating is to detect even small leukaemic populations (~10-20 leukaemic cells) among 300.000 up to 1.000.000 cells.
Goal
Provide motivated solutions to approach the problem, compare the proposed method against the state of the art. Thus, the tasks are as follows:
- Creation of training and evaluation data set: Merging all files of similar cell subsets of the first training data-set to give cumulative data “clouds”.
- Implementation of different automatic gating methods.
- Application of methods to choose the suitable algorithmic parameter as well as choosing optimal modelling parameters to train the applied classification method;
- Evaluation and comparison of the results using the different methods, as compared to expert results.
One goal is to adapt the developed method of the AutoFLOW project to AML. There, a probabilistic model is used, where each new sample is reconstructed by artificial reference samples, which are represented by Gaussian Mixture Models (GMM). The artificial samples are calculated from a training set by non-negative matrix factorization leading to a lower dimensional feature space. A Bayes decision is done to assign each observation to one of the specified sub-populations in the training set.
It should be evaluated, if the developed approach can be applied for AML.
Workflow
- Literature research on existing methods for MRD determination
- Get familiar with FCM data
- Get familiar with the current Framework for AML (R)
- Development and evaluation of the system
- Written thesis (in English) and final presentation
References
- K. Lo, R. Brinkman and R. Gottardo, Automated gating of flow cytometry data via robust model based clustering, Cytometry Part A, 73(4): 321-332.
- Bashashati and R. Brinkman, A survey of flow cytometry data analysis methods, Advances in Bioinformatics, Article ID 584603, pp.1-19, 2009.
- N. Aghaeepour, G. Finak, H. Hoos, T. R. Mosmann, R. Brinkman, R. Gottardo, and R. H. Scheuermann, Critical assessment of automated flow cytometry data analysis techniques, Nature Methods, vol. 10, no. 3, pp. 228–238, Mar. 2013.
- M. Reiter, J. Hoffmann, F. Kleber, A. Schumich, G. Peter, F. Kromp, M. Kampel and M. Dworzak, Towards automation of flow cytometric analysis for quality-assured follow-up assessment to guide curative therapy for ALL in children , In memo – magazine of european medical oncology, 7(4), pp. 219-226, Dec, 2014. Journal Article.
